Random Forest with PySpark
The dataset is wage and other data for a group of 3000 male workers in the mid-Atlantic region.
Exploration
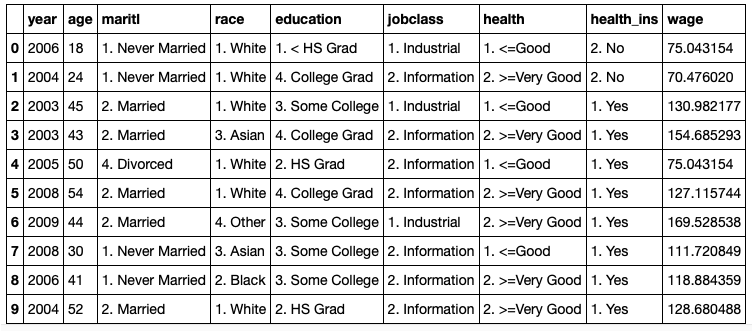
The dataset looks as follows:
Preprocessing
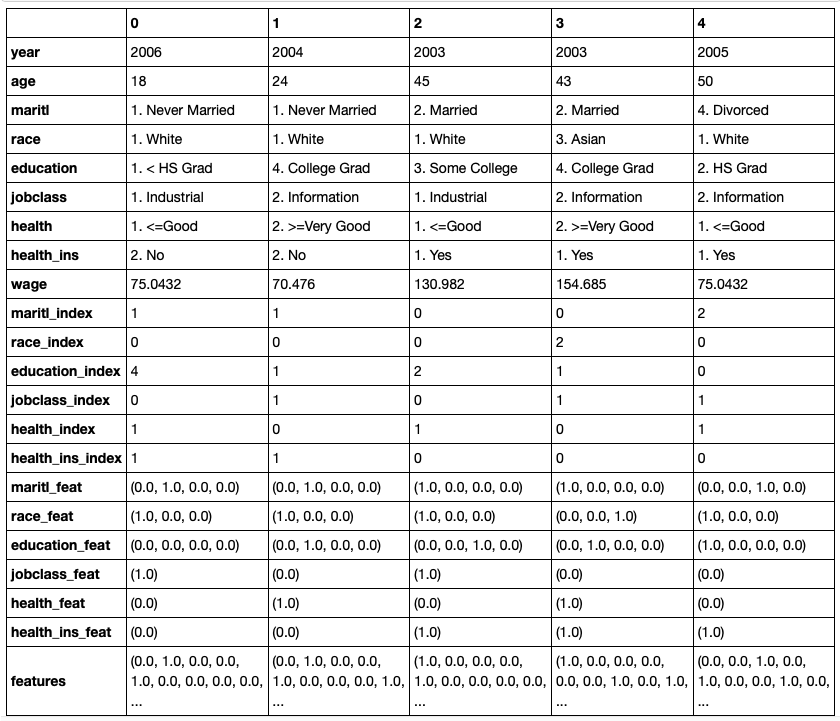
Codified the columns maritl, race, education, jobclass, health, and health_ins. The codification was a combination of a StringIndexer and a OneHotEncoder. For example, for maritl, StringIndexer created a column maritl_index, and OneHotEncoder created a column maritl_feat.
Investigated the parameters of StringIndexer so that the labels are indexed alphabetically in ascending order so that, for example, the 1st index for maritl_index corresponds to 1. Never Married, the 2nd index corresponds to 2. Married, and so forth.
Also, investigated the parameters of OneHotEncoder so that there are no columns dropped as it is usually done for dummy variables. This is, marital status should have one column for each of the classes.
The pipeline created a column features that combines year, age, and all codified columns.
Random Forest
Created three pipelines that contain three different random forest regressions that take in all features from the wage_df to predict wage. These pipelines have as first stage the pipeline created earlier fitted to the training data.
- Random forest with maxDepth=1 and numTrees=60
- Random forest with maxDepth=3 and numTrees=40
- Random forest with maxDepth=6, numTrees=20
Evaluation
Computed the RMSE of the models on validation data. The minimum value was 33.44. Random forest with maxDepth=6 and numTrees=20 performed the best on the test data.
Feature Importance
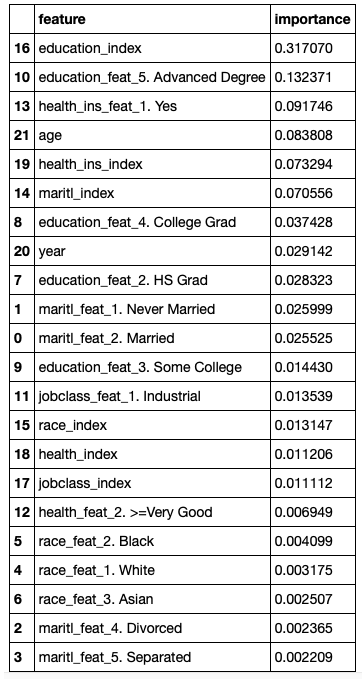
Created a pandas dataframe feature_importance with the columns feature and importance which contains the names of the features. Gave appropriate column names such as maritl_1_Never_Married. Built these feature names by using the labels from the fitted StringIndexer above. Used as feature importance as determined by the random forest of the final model (final_model). Sorted the pandas dataframe by importance in descending order and display.
Inspection
The tree looked as follows:

Conclusion

